Story
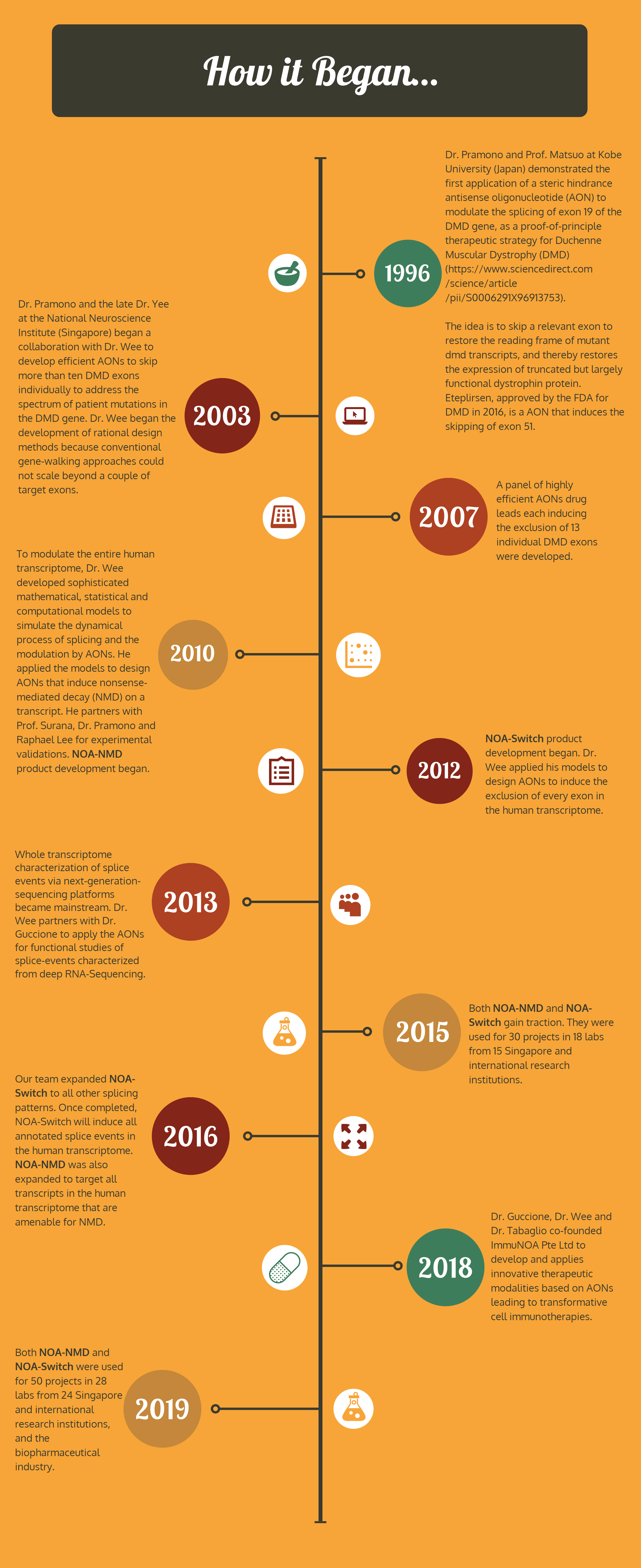

Dissecting the complexity at the transcriptomic level is requisite to comprehend the science of life and diseases. This has been impeded by current techniques for functional studies of splice-isoforms, which are non-scalable to the high-throughput generation of transcriptome data.
We hope, through efficient and specific modulation of the human transcriptome, our products will simplify the process of unravelling disease mechanisms, and catalyse the discovery and validation of novel drug targets.